| Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase |
| From: PublishDate:2012-06-26 Hits: |
Box H/ACA RNA-protein complexes (H/ACA RNPs) are ancient molecular machines conserved from archaea to eukaryotes that mediate pseudouridine synthesis, eukaryotic ribosome biogenesis and vertebrate telomere formation. Although studies of archaeal H/ACA RNP has led to a rather detailed understanding of its structure and mechanism of action, the assembly and architecture of eukaryotic H/ACA RNP remain unclear. The laboratory of Dr. Keqiong Ye, at the National Institute of Biological Science, Beijing, has reconstituted functional yeast H/ACA RNP, analyzed its assembly, activity and structure and found many eukaryotic-specific structural features of H/ACA RNP. Their results were recently published in Genes & Development on November 15, 2011. The difficulty in expression and purification of recombinant H/ACA proteins has hindered the structural and functional analysis of H/ACA RNPs for a long time. The Ye’s group has successfully obtained well soluble recombinant H/ACA proteins by utilizing a co-expression and co-purification strategy and reconstituted functional yeast box H/ACA RNPs. Taking advantage of this reconstitution system, they identified that the protein-RNA interaction occurring at the upper stem region of eukaryotic H/ACA RNA is dispensable for the activity, revealing a major difference in the functional organization between archaeal and eukaryotic complexes. Although eukaryotic H/ACA RNAs have a conserved two-hairpin structure, they found that each isolated single hairpin of yeast H/ACA RNA is able to form an independent structural and functional unit. They crystallized a ternary H/ACA protein complex of Cbf5, Nop10 and Gar1 and collected a high-quality 1.9 Å x-ray diffraction dataset at Beijing Synchrotron Radiation Facility beamline 3W1A. The structure determined by molecular replacement reveals many eukaryotic-specific features of H/ACA RNP. The core domain of yeast Gar1 contains a C-terminal extension that forms hydrophobic contacts with the thumb loop of Cbf5 (Fig. 2). Related biochemical analyses show that the extension of Gar1 regulates substrate turnover and likely facilitates Gar1 to replace the assembly factor Naf1, which binds at the same site of Cbf5 during H/ACA RNP biogenesis in vivo. Mutations in human Cbf5 (dyskerin) have been shown to lead to a rare inheritable disease dyskeratosis congenita (DC). The eukaryotic-specific N-terminal extension of dyskerin is a hot spot of DC mutation. The structure of yeast Cbf5 reveals that the extension forms an extra structural layer on the PUA domain, providing an improved model to understand the mutational effect (Fig. 3). This work using biochemical experiments and X-ray crystallographic analyses in BSRF provides novel insight into eukaryotic H/ACA RNP structure. The future work is to determine the structure of fully assembled yeast H/ACA RNP in various functional states and to understand the structural adaption of H/ACA RNP during evolution.
Figure 1. Crystal structure of the yeast Cbf5-Nop10-Gar1 complex and comparison with archaeal H/ACA RNP structure.
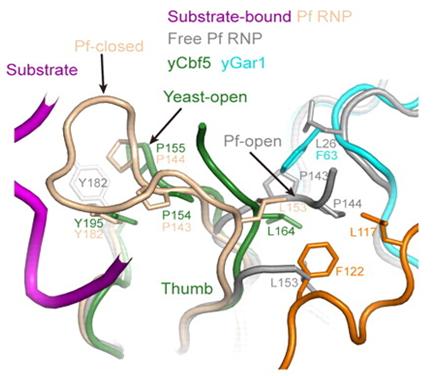
Figure 2. Yeast Gar1 possessed a conserved C-terminal extension (orange) that contacts the Cbf5 thumb loop (green) via hydrophobic interactions and regulates substrate loading and release.
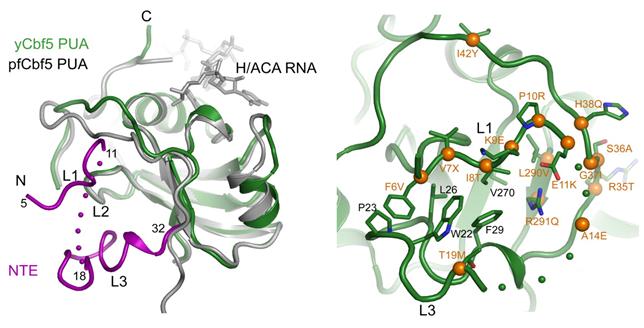
Figure 3. The N-terminal extension of yeast Cbf5 (magenta) forms an extra structural layer on the PUA domain and harbors many dyskeratosis congenita mutation sites (orange balls).
Article: Shuang Li, Jingqi Duan, Dandan Li, Bing Yang, Mengqiu Dong, and Keqiong Ye*.Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase. Genes & Development, 2011, 25, 2409-2421. |
|
|
| Chinese
Science Highlights
Home /
Copyright © 2011 - 2012 Beijing Synchrotron Radiation Facility