| SARS CoV nsp16/nsp10 protein complex: A unique ribose 2’ O-methyltransferase |
| From: PublishDate:2012-06-26 Hits: |
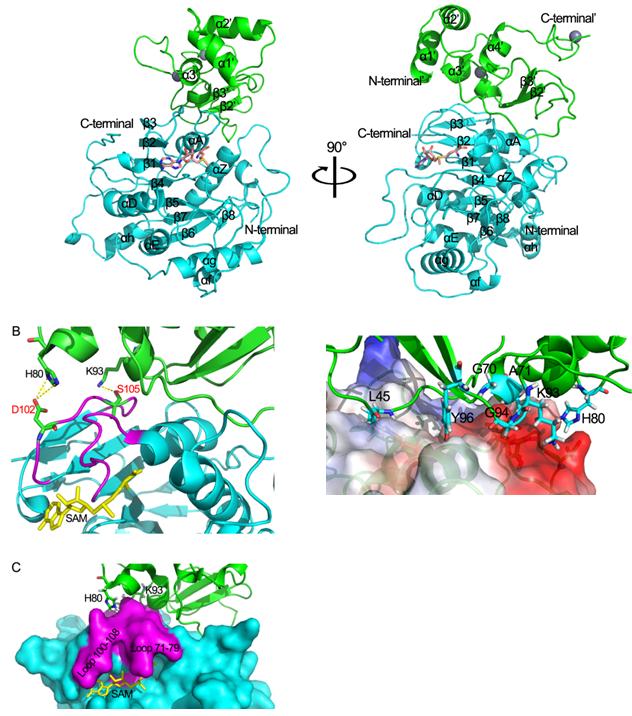
The 5′-cap structure is a distinct feature of eukaryotic mRNAs, and eukaryotic viruses generally modify the 5′-end of viral RNAs to mimic cellular mRNA structure, which is important for RNA stability, protein translation and viral immune escape. SARS coronavirus (SARS-CoV) encodes two S-adenosyl-L-methionine (SAM)-dependent methyltransferases (MTase) which sequentially methylate the RNA cap at guanosine-N7 and ribose 2′-O positions, catalyzed by nsp14 N7-MTase and nsp16 2′-O-MTase, respectively. A unique feature for SARS-CoV is that nsp16 requires non-structural protein nsp10 as a stimulatory factor to execute its MTase activity. Yet a team composed of researcher from Wuhan University and Nankai University has gained insight into the mysterious structure and biochemical feature of nsp16/nsp10 protein complex. Their research has been published on October 13th, 2011 in Plos Pathogens. Previous work of this team indicate that nsp10 may function as a stimulatory factor for nsp16 and is required for the 2′-O-MTase activity of nsp16. Now they proved that nsp10 assists nsp16 to bind substrate for its methyltransferase activity, include capped RNA substrate and the methyl donor S-adenosyl-L-methionine (SAM), also they present structure of nsp16/nsp10 complex. They show that nsp16 possesses the characteristic fold of the class I MTase family, comprising a seven-stranded β-sheet surrounded by α-helices and loops. But differences between nsp16 and other methyltransferase which can exert methyltransferase alone, is that nsp16 possesses a shorter αD helix. This feature makes the SAM binding cleft of nsp16 seems more flexible and lower its SAM binding activity, this can explain why nsp16 needs another protein to promote its activity. They also find one hydrogen bond forms between Lys-93 of nsp10 and Ser-105 of nsp16, and two salt bridges exist between His-80 ND1 and NE2 from nsp10 with Asp-102 OD2 from nsp16, these interactions sustain one wall of SAM binding cleft of nsp16, and thus promote the SAM binding activity of nsp16. SARS-CoV nsp16 is the only 2′-O-MTase currently known that needs a stimulatory subunit for exerting its methyltransferase activity. These mechanisms could be explained based on the crystal structure of nsp16/nsp10 complex, and confirmed by mutational analysis. mRNA capping and methylation play important roles in mRNA stability, processing, transport and protein translation and thus The MTase active site has been suggested as a drug target for developing antiviral drugs, However, the MTase fold is structurally conserved between viral and cellular MTases, and it is thus difficult to obtain antiviral compounds with high specificity targeting MTase active sites. For this reason, it looks more promising to target the interface of nsp16 and nsp10, which is unique to coronaviruses.
Structure of nsp16/nsp10 protein complex, determined at 2.0 Å resolution using synchrotron radiation at BSRF.
Article: Yu Chen, Ceyang Su, Min Ke, Xu Jin, Lirong Xu, Zhou Zhang, Andong Wu, Ying Sun, Zhouning Yang, Po Tien, Tero Ahola, Yi Liang, Xinqi Liu*, Deyin Guo*, Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2’-O-Methylation by nsp16/nsp10 Protein Complex. PLoS Pathog. 2011, 7(10), e1002294. |
|
|
| Chinese
Science Highlights
Home /
Copyright © 2011 - 2012 Beijing Synchrotron Radiation Facility