| Structural insights into the function of ribosomal RNA methyltransferases RlmG and RsmE |
| From: PublishDate:2013-06-15 Hits: |
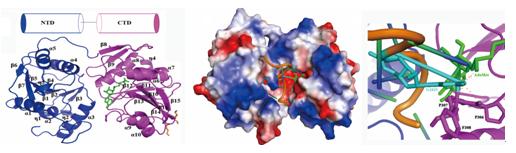
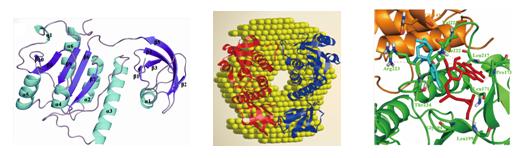
Methylation of ribosomal RNA (rRNA) nucleotides plays an important role in the biogenesis and regulation of the ribosome in all living organisms. This modification is closely associated with fine-tuning the process of protein synthesis and bacterial resistance to antibiotics. A team from Center for Protein Structure and Function, Institute of High Energy Physics, CAS, has gained insight into the structure and function of ribosomal RNA methyltransferases (MTases) RlmG (m2G1835) and RsmE (m3U1498) to reveal the novel catalytic mechanisms. Their research has been published in RNA and Journal of Molecular Biology of 2012, respectively. RlmG is a specific AdoMet-dependent MTase responsible for N2-methylation of G1835 in 23S rRNA of Escherichia coli. Single wavelength anomalous data (SAD) from seleno-RlmG crystals were collected on the beamline station 3W1A of Beijing Synchrotron Radiation Facility (BSRF). The structure of RlmG is composed of two homologous domains: N-terminal domain (NTD) in the recognition and binding of the substrate, and C-terminal domain (CTD) in AdoMet-binding and catalytic process.RNA-binding properties of NTD and CTD characterized by both gel electrophoresis mobility shift assays and isothermal titration calorimetry showed NTD could bind RNA independently and RNA binding was achieved by NTDaccomplished by coordinating role of CTD.The model of RlmG-AdoMet-RNA complex suggested that RlmG may unfold its substrate RNA in the positively charged cleft between NTD and CTD, and then G1835 disengages from its Watson-Crick pairing with C1905 and flips out to insert into the active site. The structure and biochemical studies provide novel insights into the catalytic mechanism of G1835 methylation. RsmE is the founding member of a new RNAMTase family responsible for methylation of U1498 in 16S rRNA in Escherichia coli. Diffraction data of RsmE were collected on the beamline station 1W2B of BSRF.The crystal structure in monomer showed that it consists of two distinct but structurally related domains: the PUA-like RNA recognition and binding domain and the conserved MTase domain with a deep trefoil knot.Analysis of small-angle X-ray scattering (SAXS) data revealed RsmE forms a flexibledimeric conformationwhich may be essential for substrate-binding.The AdoMet-binding characteristic determined by isothermal titration calorimetry suggested there is only one AdoMet molecule bound in the subunit of the homodimer. In vitromethylation assay of the mutants based onthe RsmE-AdoMet-UMP complex model showed key residues involved in substrate-binding and catalysis. The MTase domain of one subunit in dimeric RsmE is responsible for binding of one AdoMet molecule and catalytic process while the PUA-like domain in the other subunit is mainly responsible for recognition of one substrate molecule (the rRNA fragment and ribosomal protein complex). The methylation process is required by collaboration of both subunits and dimerization is functionally critical for catalysis.In general, this study provides new information on the structure-function relationship of RsmE, and thereby suggests a novel catalytic mechanism.
Single wavelength anomalous data (SAD) from seleno-RlmG crystals were collected on the beamline station 3W1A of BSRF and the RlmG-AdoMet comolex structure was determined at 2.25 Å resolution. RlmG is composed of two homologous domains: N-terminal domain (NTD) and C-terminal domain (CTD). The model of RlmG-AdoMet-RNA complex suggested that RlmG may unfold its substrate RNA in the positively charged cleft between NTD and CTD, and then G1835 disengages from its Watson-Crick pairing with C1905 and flips out to insert into the active site. Diffraction data of RsmE were collected on the beamline station 1W2B of BSRF. The crystal structure showed that it consists of two distinct but structurally related domain.Analysis of SAXS data revealed RsmE forms a flexibledimeric conformationwhich may be essential for substrate-binding.Only one AdoMet molecule bound in the subunit of the homodimer. Inthe RsmE-AdoMet-UMP complex model, the MTase domain of one subunit is responsible for binding of one AdoMet molecule and catalytic process while the PUA-like domain in the other subunit is mainly responsible for recognition of one substrate molecule.
Article: 1、hang H, Gao Z, Wei Y, Wang W, Liu G, Shtykova E, Xu J and Dong Y. (2012) Structural insights into the function of 23S rRNA methyltransferase RlmG (m2G1835) from Escherichia coli. RNA. 18: 1500-1509. 2、Zhang H, Wan H, Gao Z, Wei Y, Wang W, Liu G, Shtykova E, Xu J and Dong Y. (2012) Insights into the Catalytic Mechanism of 16S rRNA Methyltransferase RsmE (m3U1498) from Crystal and Solution Structures. Journal of Molecular Biology. 423: 576-589. |
|
|
| Chinese
Science Highlights
Home /
Copyright © 2011 - 2012 Beijing Synchrotron Radiation Facility